Affymetrix GeneChip Microarray
We offer a complete microarray service for gene expression and miRNA analysis. We offer the Affymetrix Clariom D & S arrays for gene expression. The Clariom S array is utilized for differential gene expression and pathway analysis. The Clariom D arrays provide transcriptome-wide gene- and exon- level expression profiles. The miRNA v4.0 array is a powerful tool for studying the role of small non-coding RNAs.
The microarry service includes sample labeling, hybridization, scanning and preliminary data analysis.
Clariom D arrays provide a highly detailed view of the transcriptome and allow translational research scientists to generate biomarker signatures quickly and easily. The Clariom D arrays provides the most intricate transcriptome-wide gene- and exon-level expression profiles, including the ability to detect alternative splicing events of coding and long non-coding (lnc)RNA. It is comparable to performing RNASeq analysis.
Clariom S arrays provide a gene-level view of the transcriptome and focus on well-annotated genes, providing researchers with the ability to perform gene-level expression profiling studies and to quickly assess changes in key genes and pathways. The Clariom S array design provides extensive coverage of all known well-annotated genes.
miRNA v4.0 array is a powerful tool for studying the role of small non-coding RNAs. The miRNA 4.0 array is designed to interrogate all mature miRNA sequences in miRBase Release 20 design.
It is estimated that more than 30% of protein translation of coding genes is regulated by miRNA. There is also a large amount of growing evidence suggesting miRNA interacts with long non-coding RNA in the signaling networks that regulate alternative splicing events, which impacts cellular processes such as apoptosis, proliferation, and differentiation– all of which have shown to be causative elements in diseases such as cancer. Measuring the changes in these critical nodes of regulation is extremely important for deciphering the biological context of differentially expressed genes.
Sample Requirements
• 150 ng high quality total RNA
• Minimum Concentration of 50 ng/µl
• OD260/280 ratio > 1.8
• Bioanalyzer analysis to assess the RNA quality is highly recommended.
Data Analysis
The core will provide preliminary data analysis using the Affymetrix Transcriptome Analysis Console (TAC) Software.
If you would like to perform additional analysis and fully utilize the power of the TAC software it is free for download. (Click here)

aCGH
We use a whole human genome tiling path BAC array for copy number analysis. This array has an effective resolution of 150kb and spans 93% of the human euchromatic genome. In addition to the basic tools/reagents, we have sophisticated bioinformatics tools at our disposal, as well as a large database of copy number variation in normals (currently over 800 individuals). We also offer genome wide copy number analysis using any of the major platforms, including NimbleGen and Agilent oligonucleotide based microarrays.
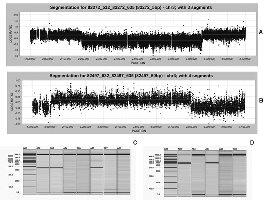
Fine tiling NimbleGen oligonucleotide array used to map chromosome 3 breakpoints with enough resolution to generate junction fragments by PCR (Jasmin Roohi, Cristina Montagna, David H. Tegay, Lance E. Palmer, John C. Pomeroy, Susan L. Christian, Norma Nowak and Eli Hatchwell. Disruption of Contactin 4 in 3 Subjects with Autism Spectrum Disorder. Journal of Medical Genetics (Rapid online publication - March 18th 2008).
